May 21, 2025
Welcome to our Google Summer of Code scholars for 2025!
November 30, 2024
OpenWorm Annual Meeting 2024 (DevoWorm update)
Here are the slides for the DevoWorm group's report to the OpenWorm Annual Meeting (2024). You can watch Bradly Alicea present the talk on YouTube.
Aside from all the great stuff going on in DevoWorm, there are two new tools being developed by Padraig Gleeson's team at University College London. First up is the Connectome Toolbox, which brings together multiple key datasets and visualization tools for C. elegans connectome analysis. The other is OpenWorm.ai, which uses a large language model (LLM) hosted on Huggingface to query information about C. elegans biology.
Here are some slides from the presentation. Click to enlarge. Thanks to Mehul Arora and Pakhi Banchalia for their Google Summer of Code efforts.
May 22, 2024
Google Summer of Code 2024
Welcome to the new Google Summer of Code scholars for 2024! INCF is sponsoring four students for which I (Bradly Alicea) am acting as mentor: two students for the DevoWorm group (via the OpenWorm Foundation community) and two students for the Orthogonal Research and Education Laboratory.
D-GNNs (sponsored by the DevoWorm group)
Congratulations to Pakhi Banchalia and Mehul Arora for being accepted to work on the Developmental Graph Neural Networks (D-GNNs) project. Pakhi will be working on incorporating Neural Developmental Programs (NDPs) into GNN models. Mehul will be working on hypergraph techniques for developmental lineage trees and embryogenesis [1]. Himanshu Chougule (Google Summer of Code scholar) is a co-mentor for this project.
Congratulations to Sarrah Bastawala for being accepted to work on the Open-source Sustainability project [2]. Sarrah is incorporating Large Language Models (LLMs) into the collection of agent-based approaches for this project. Jesse Parent is a co-mentor for this project.
We also have two Open-source Development scholars for Summer 2024: Adama Koita and Shubham Soni. They will be participating in the Orthogonal Lab's open-source weekly meetings in addition to projects based around Virtual Reality and Open-source Sustainability, respectively.
November 30, 2023
OpenWorm Annual Meeting 2023 (DevoWorm update)
It's that time again -- time for the OpenWorm Annual meeting. Below are the slides I presented on progress and the latest activities in the DevoWorm group. If anything looks interesting to you, and you would like to contribute, please let me know. Click on any slide to enlarge.
May 8, 2023
Google Summer of Code 2023
Welcome to the new Google Summer of Code scholars for 2023! INCF is sponsoring four students for which I (Bradly Alicea) am acting as mentor: two students for the DevoWorm group (via the OpenWorm Foundation community) and two students for the Orthogonal Research and Education Laboratory. These four students are pursuing three projects.
D-GNNs and DevoLearn (sponsored by the DevoWorm group)
Congratulations to Himanshu Chougule and Sushmanth Reddy Mereddy for being accepted to work on the Developmental Graph Neural Networks (D-GNNs) project. Sushmanth has been contributing to the DevoLearn platform for quite some time and will spend this summer working on improving the image segmentation to GNN embedding pipeline. Himanshu was a part of last Summer's GSoC cohort in the Orthogonal Research and Education Laboratory, working on the Open-source Sustainability project. His work resulted in developing Agent-based Model-Reinforcement Learning hybrid models. This year, he will be working on building Topological Data Analytic capabilities into the D-GNN pipeline.
Mayukh Deb and Jiahang Li are co-mentors for this project.
Virtual Reality for Research and Open-source Sustainability (sponsored by the Orthogonal Research and Education Lab)
Congratulations to Vrushali Nandurkar and R.V. Rajagopalan for being accepted to work on the projects Virtual Reality for Research and Open-source Sustainability, respectively. Vrushali is enthusiastic about working on creating open-source virtual world assets for scientific research and educational initiatives. R.V. will be working on a continuation of the Open-source Sustainability project, helping to augment the existing models and web interface.
Jesse Parent is a co-mentor for both projects, while Brian McCorkle is a co-mentor for the Open-source Sustainability project.
March 3, 2023
Ancient Embryogenesis and Evolutionary Origins
"Darwin as an Embryo". In this case, Neoteny really does recapitulates Phylogeny! COURTESY: Stable Diffusion.
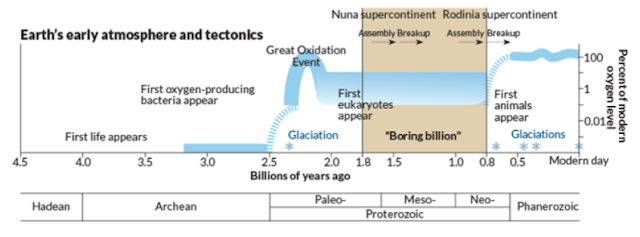
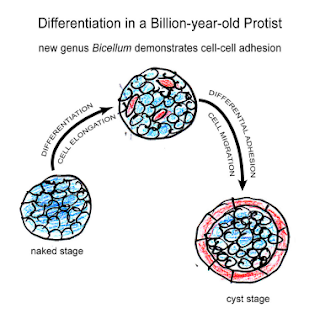
For this year's delayed Darwin Day post, I will present some of the latest work on ancient embryos which we have been discussing in the DevoWorm group meetings. While this is by no means a complete review, we will discuss the earliest fossil evidence for eggs, embryos, and nervous systems (in animals, not plants), in addition to the conditions that lead to their emergence. In short, how did we get to embryos from a universal common ancestor with bacteria and archebacteria, and why do only different types of Eukaryotes (plants, radial symmetrical Metazoans, and bilateral Metazoans) have embryos?
Tree of Life (genome tree) from Hug et.al [1] with three domains. Click to enlarge.
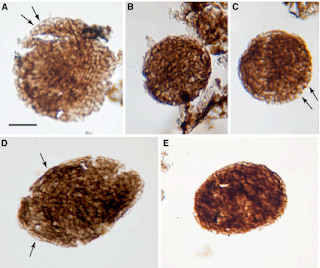
We begin in the Cambrian, where the firs bilaterian appeared around 600 million years ago, approximately 70-80 million years before the Cambrian explosion [2]. In between the emergence of bilaterians, several key innovations occurred that suggests the origins of embryos and egg-laying. The first are the presence of fossilized burrows [2] for egg-laying behaviors. By the end of the Cambrian explosion, early pancrustecean arthropod species were possibly subject to life-history tradeoffs related to clutch size [3]. Another key innovation is direct evidence in the form of well-preserved multicellular structures from the period leading up to the Cambrian explosion that show a transition in cell geometry from a 2-cell stage to a cleavage stage [4]. As representative of a variety of ancestral algae species from the Doushantuo formation, these remains have not been connected to any particular adult form. However, they do demonstrate oogenesis and cleavage [2]. Finally, the functional genomics of developmental pattern formation emerged during this time [5]. This includes a ProtoHox cluster in ancestral cnidarians [6], Hox gene duplication [7], and an increase in body size and shape diversity alongside the advent of bilaterian bauplans [8]. Multiple Hox gene families may have served the role of promoting directed locomotion that in turn promoted active exploration of the environment [7].
Images of a potential early embryo, including the 2-cell and cleavage stages [from 4]. Click to enlarge.
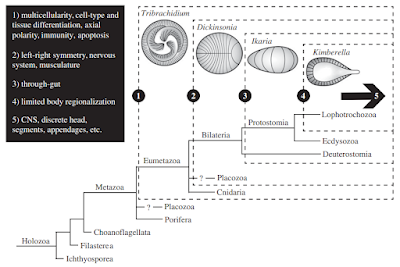
The Ediacaran (630-540 million years ago) has yielded a large number of potential embryonic forms. In the Ediacaran biota, we find a number of Metazoan remains with no clear phylogenetic position. However, Evans et.al [9] propose that early embryos evolved independently (with several origins) in the bilaterian clade. However, during this time, a number of general trends emerge that enabled modern bilaterian adult forms. As previously discussed, Multicellular structures with distinct cell types, axial polarity, and anatomical segmentation [10, 11] emerged during this time. Left-right symmetry was a related feature of these embryos [11]. So-called polarized elements [12] such as microtubules, flagella, and apical-to-basal orientation were all found soon after the last Eukaryotic common ancestor (LECA). The evolution and diversification of polarity proteins is consistent with this timeline [12]. Other organismal structures such as a gut, sub-specialization of the phenotype, and a nervous system with heads and appendages are also features of note. We will talk about the emergence of nervous systems later on.
Scenario for the origins of development in bilaterians from [9]. Click to enlarge.
June 15, 2022
Google Summer of Code 2022 in the OpenWorm Community (DevoWorm)
This year, we were able to fund two students per project. They will be working on complementary solutions to each problem, and we will see how far they get by the end of the Summer.
D-GNNs (Developmental Graph Neural Networks)
The description for this project is as follows:
Biological development features many different types of networks: neural connectomes, gene regulatory networks, interactome networks, and anatomical networks. Using cell tracking and high-resolution microscopy, we can reconstruct the origins of these networks in the early embryo. Building on our group's past work in deep learning and pre-trained models, we look to apply graph neural networks (GNNs) to developmental biological analysis.
The contributor will create graph embeddings that resemble actual biological networks found throughout development. Potential activities include growing graph embeddings using biological rules, differentiation of nodes in the network, and GNNs that generate different types of movement output based on movement seen in microscopy movies. The goal is to create a library of GNNs that can simulate developmental processes by analyzing time-series microscopy data.
When completed, D-GNNs will become part of the DevoWorm AI library. Ultimately, we will be integrating the GNN work with the DevoLearn (open-source pre-trained deep learning) software.
Jiahang Li is a first year MPhil candidate in Computing Department at Hong Kong Polytechnic University. His research interests cover graph representation learning and its applications. Jiahang's approach to the project is to provide a pipeline that converts microscopic video data of C. elegans and other organisms into graph structures, on which advanced network analysis techniques and graph neural networks will be employed to obtain high-level representation of embryogenesis and to solve applied problems.
Wataru is a student at Kyoto University with interests in Machine Learning (in particular Graph Neural Networks) and Neuroimaging.
Digital Microspheres
The description for this problem is as follows:
This project will build upon the specialized microscopy techniques to develop a shell composed of projected microscopy images, arranged to represent the full external surface of a sphere. This will allow us to create an atlas of the embryo’s outer surface, which in some species (e.g. Axolotl) enables us to have a novel perspective on neural development.
The contributor will build a computational tool that allows us to visualize 4D data derived from the surface of an Axolotl embryo. The spatial model and animation (4th dimension) of microscopy image data can be created in a 3-D modeling software of your choice.
This project is based on previous research by DevoWorm contributors Richard Gordon and Susan Crawford-Young. The flipping and ball microscopy research involve the design and fabrication of specialized microscopes to image embryos in a 4-D context (3 dimensions of space plus time).
Spherical Embryo Maps: Gordon, R. (2009). Google Embryo for Building Quantitative Understanding of an Embryo As It Builds Itself. II. Progress Toward an Embryo Surface Microscope. Biological Theory, 4, 396–412.
Flipping Microscopy: Crawford-Young, S., Dittapongpitch, S., Gordon, R., and Harrington, K. (2018). Acquisition and reconstruction of 4D surfaces of axolotl embryos with the flipping stage robotic microscope. Biosystems, 173, 214-220.
Ball Microscopy: Crawford-Young, S.J. and Young Williment, J.L. (2021). A ball microscope for viewing the entire surface of amphibian embryos. Biosystems, 208, 104498.
Karan Lohaan
Karan is a student at Amrita Vishwa Vidyapeetham University, and is a member of the AMFoss program there. He is interested in Machine Learning and Image Processing.
Harikrishna Pillai
I am Harikrishna pursuing my B.Tech in Computer Science and Artificial Intelligence from Amrita Vishwa Vidyapeetham University. I completed my schooling in Mumbai. I started with python as my first language and eventually developed interest for AI. Due to my interest in Android apps, I have done Android development in Kotlin. Also, I have been interested in open source for some time now and therefore, I wanted to start my open source journey with GSoC.
We also have two GSoC mentors for these projects: Bradly Alicea is a mentor for D-GNNs and Digital Microspheres, and Jesse Parent is a mentor for D-GNNs. Richard Gordon and Susan Crawford-Young are serving as collaborators for the Digital Microspheres project.
If you would like to check on their progress, please check out our weekly meetings available on our YouTube channel.



.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)



.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)
.png)